First of all, I’d like to thank my readers for the lots of feedback on my last post on beautiful outputs in R. I tried to consider all suggestions, updated the existing table-output-functions and added some new ones, which will be described in this post. The updated package is already available on CRAN.
This posting is divided in two major parts:
- the new functions are described, and
- the new features of all table-output-functions are introduced (including knitr-integration and office-import)
New functions
First I want to give an overview of the new functions. As you may have noticed, all table-output-functions have new parameters, which enable you to modify the appearance and retrieve objects for knitr-integration and so on. This is described below.
Viewing imported SPSS data sets
As I have mentioned some times before, one purpose of this package is to make it easier for (former) SPSS users to switch to and use R. Beside the data import functions (see all functions beginning with sji) I now added two functions, where one is specifically useful for SPSS data sets, while the other one is generally useful for data frames.
With the function sji.viewSPSS you can easily create a kind of „code plan“ for your data sets. Note that this function only works for SPSS data sets that have been imported using the sji.SPSS function (because else variable and value label attributes are missing)! The function call is quite simple. Load the library with require(sjPlot) and run the following example:
data(efc) sji.viewSPSS(efc)
This will give you an overview of: Variable number, variable name, variable label, variable values and value labels:

You can suppress the output of values and value labels if you just want to quickly inspect the variable names. The table can also be sorted either by variable number or by variable name.
Description and content of data frames
If you want to inspect the data frame’s variables, you can use the sjt.df function. By default, this function calls the describe-function from the psych-package and prints the output as HTML-table:
data(efc) sjt.df(efc)

If you set the parameter describe=FALSE, you can view the data frame’s content instead. See this example, where alternate row colors are activated and the table is ordered by column „e42dep“:
sjt.df(efc[1:20,1:5], alternateRowColors=TRUE,
orderColumn="e42dep", describe=FALSE)

Be careful when applying this function to large data frames, because it becomes very slow then…
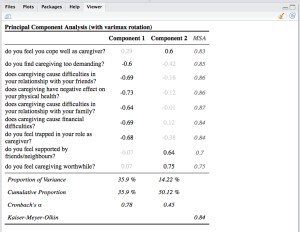
Principal Component Analysis and Correlations
Two more new functions are sjt.pca for printing results of principal component analyses and sjt.corr for printing correlations. Printing PCA results will give you an overview of all extracted factors, where the highest factor loading is printed in black, while the other factor loadings are a bit faded (thus, it’s easier to see which item belongs to which factor). Furthermore, you can print the MSA for each item, the Cronbach’s Alpha value for each „scale“ and other statistics:
data(efc) # retrieve variable and value labels varlabs <- sji.getVariableLabels(efc) # recveive first item of COPE-index scale start <- which(colnames(efc)=="c82cop1") # recveive last item of COPE-index scale end <- which(colnames(efc)=="c90cop9") # create data frame with COPE-index scale df <- as.data.frame(efc[,c(start:end)]) colnames(df) <- varlabs[c(start:end)] sjt.pca(df, showMSA=TRUE, showVariance=TRUE)
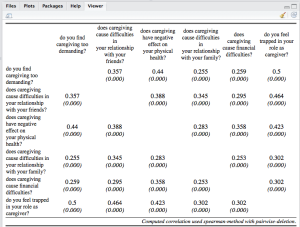
The next example is a correlation table. Note: This table may look more beautiful if opened in a web browser (because of more space). And second note: See the usage of the CSS-parameter! (more on this later)
sjt.corr(df, pvaluesAsNumbers=TRUE,
CSS=list(css.thead="border-top:double black; font-weight:normal; font-size:0.9em;",
css.firsttablecol="font-weight:normal; font-size:0.9em;"))
Stacked frequencies and Likert scales
The last new table-output-function is sjt.stackfrq, which prints stacked frequencies of (Likert) scales.
data(efc)
# recveive first item of COPE-index scale
start <- which(colnames(efc)=="c82cop1")
# recveive first item of COPE-index scale
end <- which(colnames(efc)=="c90cop9")
# retrieve variable and value labels
varlabs <- sji.getVariableLabels(efc)
vallabs <- sji.getValueLabels(efc)
sjt.stackfrq(efc[,c(start:end)],
valuelabels=vallabs['c82cop1'],
varlabels=varlabs[c(start:end)],
alternateRowColors=TRUE)

Similar to the sjp.stackfrq function (see this posting), you can order the items according to their lowest / highest first value etc.
Tweaking the table-output-functions and integrating output into knitr
In this section, important new parameters of the table-output-functions are described.
Each sjt function as well as sji.viewSPSS now have following parameters:
- CSS
- useViewer
- no.output
And all of them (invisibly) return at least following values:
- the web page style sheet (
page.style), - the web page content (
page.content), - the complete html-output (
output.complete) and - the html-table with inline-css for use with knitr (
knitr)
Parameters explained
CSS
The table-output is in HTML format, using cascading style sheets to modify the appearance of tables. You can inspect the page.style and page.content parameters to see which CSS classes are used in the HTML-table, for instance:
> value <- sjt.df(efc)
> value$page.style
[1] "<style>\ntable { border-collapse:collapse; border:none; }\ncaption { font-weight: bold; text-align:left; }\n.thead { border-top: double; text-align:center; font-style:italic; font-weight:normal; padding:0.2cm; }\n.tdata { padding:0.2cm; text-align:left; vertical-align:top; }\n.arc { background-color:#eaeaea; }\n.lasttablerow { border-top:1px solid; border-bottom: double; }\n.firsttablerow { border-bottom:1px solid; }\n.leftalign { text-align:left; }\n.centertalign { text-align:center; }\n.firsttablecol { }\n.comment { font-style:italic; border-top:double black; text-align:right; }\n</style>"
To use the CSS parameter, you must define a list with values, where the value-name equals the css-class-name with css. prefix. If you want to change the appearance of the first table column (with variable names), use:
sjt.df(efc, CSS=list(css.firsttablecol="color:blue;font-style:italic;"))
Note that each style-definition in the parameter-list has to end with ;, because sometimes style-attributes are concatenated and thus need this separator char. Refer to the function-help to see more examples…
useViewer and no.output
With useViewer set to FALSE, you can simply force opening the html-table-output in a web browser, even if a viewer is available. With no.output set to TRUE, you can suppress the table output completely. This is useful if you want to integrate the tables in your knitr-documents…
Knitr integration
As said above, each sjt-function returns an object where you can access the created html-output. The $knitr object contains the pure html-table (without HTML-pageheader or body-tags) with inline CSS (thus, no class-attributes are used). This allows the simple integration into knitr-documents. Use following code snippet in your knitr-documents and knit it to HTML:
`r sjt.df(efc, no.output=TRUE)$knitr`
Office import improvements
When setting the file parameter, the table-ouput is saved to a file. This can be opened via MS Word, LibreOffice Writer etc. The import has been improved, so the imported table should render properly now.
Last Words…
Well, enough said. 😉 All feature available in the latest sjPlot-package.

Awesome work! Thank you very much..
Awesome work! Can we also get LATEX output for using it in .Rnw files ?
Thank you for your feedback! I originally focused on HTML because 1) I wanted to use the viewer pane as alternative view to the console output and 2) HTML-tables can be easily imported into Office apps, while LaTex / PDF cannot be easily imported. Supporting LaTex would mean quite some work and I would also have to become more familiar with LaTex syntax to properly implement LaTex support. I’m afraid and sorry to say that I’m not able to do this in the near future.
Thank you for your answer. Could you please help on how to use a different alternate row color than light gray ?
Yes, you can use the CSS parameter:
CSS=list(css.arc="background-color:#993333;")(don’t forget the ; at the end). arc stands for alternate row color. Or you can turn it completely off with the related parameter (alternateRowColor, i’m not sure about the spelling, because i’m out of Office to check).You could copy the table from HTML to excel, re-format it, and then use the Excel2LaTeX plugin to get the latex code. A bit tedious, but it works
Once again, brilliant stuff.. Thank you so much for your work and release of great tools!
Thanks for your feedback, I’m glad to hear that. 🙂
This is great. I’ve always wondered how to get things to show up in the viewer. This makes copy paste into excel really easy as well. Are you on github?
Thanks for your feedback! Yes‘ I have a github-account: https://github.com/dluedecke
But I don’t know how to integrate it in RStudio, so my project will be hosted on github (see „Adding version control to a project“ at http://www.rstudio.com/ide/docs/version_control/overview). I have installed a github.app and already set some options (https://strengejacke.files.wordpress.com/2014/03/rstudio_options.png), but when I want to perform the 3rd step (Change the version control system from (None) to Git), I have not choice for Git, just „none“…
Hat dies auf New Hampshire R Users Group (NH UseRs) rebloggt und kommentierte:
Nice post on making nice tables in R, which is generally more of a challenge than making pretty figures.
Hi guy, it is a really good job. Congrats! I have one doubt about how to use sjt.df() integrated to knitr chunk multiline, inside a for statement. Can you help me?
I’m sorry, but I haven’t worked much with knitr yet, so I’m afraid I can’t help you. Perhaps you can ask experienced knitr-users?
Decimal point alignment in tables would be a nice feature.
Great work.
Your tips for knitting tables were awesome. Do you have any advice for knitting the graphs?
I’m not sure what you mean, could you specify your request? You can use any plot output in knitr, just like I did for my RPubs documentation. See knitr demos for some examples.
I published my RPubs examples, which use plots/graphics in knitr, on GitHub.
Extremely good work. Keep it up.
Thanks a lot for your package! Coming from SPSS I nearly gave in looking for smart looking tables in R when I found your programming-work. As I’m not really familiar with programming your documentation on http://rpubs.com/sjPlot was especially helpful for me. Elsewise I’m often confused about the programmer’s terminology. But your work encourages me to try a little bit harder.
There is one thing I’d like to suggest, as I’m used to it from SPSS. I hope you’ll help me with descriptive tables (CTABLES-command).
I understand that sjt.df {sjPlot} uses describe {psych}. For my purposes it would be great if it would use something as describeBy {psych} and would allow to select the shown statistics (as flexible as your sjt.xtab {sjPlot}).
I’ve seen that you already created sjt.grpmean {sjPlot}, which is nearly what I need. But if I’m right it currently allows for one grouping variable only and it is not possible to decide on the descriptive statistics. I’d need M, (SD) and Valid-n for totals and the cells of two or even more grouping variables (and ideally shown for serveral dependent variables).
I’d be very grateful if you could help me.
Thomas
Thomas, thanks for your feedback!
The documentation on RPubs is out of date. The current online-manual is at strengejacke.de/sjPlot. These are also newer and updated to the latest package version.
To get what you would like to have (describeBy), you have to write your own workaround. I suggest you start with looking at
dplyrandtidyr, two very helpful (and basic) packages. There’s a great cheat sheet at RStudio and also a very great presentation here.describeBy from psych-package returns a list of data frames. Combine all data frames of this list via dplyr::bind_rows. Select those statistics (data frame columns) of your interest with dplyr::select. The resulting, single data frame can be printed as table with
sjt.df(data.frame, describe = F).Drop me a mail if you have further questions…
Daniel, thanks a lot for your hints and the links.
I’ve tried a while and it seems that I’ll need some more time and practice to get the totals to exactly the place I want them to be (subtotals below respectively right of the cellstatistics and table total on lower right corner). But I understand that dplyr is a mighty tool, which I wouldn’t even have looked for without your help.
Unfortunately, I’ve to stop for now. I’ll tell, if there’s some progress next week. If I stuck, I’ll make use of your kind proposal to ask once more…
Hello Daniel, after some instructional hassels it seems that I’m on a good way. I’ve nearly got what I need. Not yet as flexible as I would like it to be, but maybe with a bit more practice I’ll get something worthwhile to report here.
Unfortunately I stagnate because it seems to me that sjt.df overwrites the output-file by default. Is there a way to append new output to an existing file? It would be very helpful as I have to report a lot of different descriptive tables with varying titles and comment strings. Can you help me again?
One way is to create the tables and save their output to a new object (see return values), and then write all tables into one html-file. There’s an example in
?sjt.lm, see connecting two html-tables.Thanks again, Daniel!
The example was very helpful and I managed to get the desired html-file. For other beginners it may be helpful to void one dead end I ran into. So I report, what I’ve done:
# Aims at writing different descriptive tables into one html-file.
# There are different types of tables:
# part1, part2 and part4 are results of sjt.df
# part3 is result of sjt.xtab
# This doesn’t work – seemingly just 2 html-files are aloud
write(sprintf(„%s%s%s“,
part1$page.style,
part1$page.content ,
part2$page.content ,
part3$page.style ,
part3$page.content ,
part4$page.style ,
part4$page.content),
file = „C:/Test/test.html“)
# This works, but change of styles is not possible.
# The last style is used, when there are identical style-names.
write(sprintf(„%s%s%s“,
part1$page.style,
part1$page.content ,
part2$page.content ),
file = „C:/Test/test.html“)
write(c(part3$page.style , part3$page.content , part4$page.style , part4$page.content) ,
file = „C:/Test/test.html“ ,
append=TRUE)
Hello Daniel,
it needed some time and a lot of help of some colleagues (thanks to Carla, Thomas and Michael). But now it seems to work.
Following is the code for a n by k table that show several statistics. I hope it is useful to others.
Thanks for your help
Thomas
#########begin of code
#needed packages####
library(„sjPlot“)
library(gdata)
#definition of the function that calculates the table####
createFunctionCrossMatrix <- function (AV,
UV1 = NULL,
UV2 = NULL,
stats = NULL,
statnames = NULL)
{
AVWithoutNA <- AV[!(is.na(UV2) | is.na(UV1))]
UV1WithoutNA <- to_label(UV1)[!(is.na(UV2) | is.na(UV1))]
UV2WithoutNA <- to_label(UV2)[!(is.na(UV2) | is.na(UV1))]
allresults <- list("matrix", length(stats))
for (i in 1:length(stats)) {
stat = match.fun(stats[[i]])
statname = statnames[i]
subresult <- tapply(AVWithoutNA, INDEX=list(UV1WithoutNA, UV2WithoutNA), FUN=stat)
uv2total <- tapply(AVWithoutNA, INDEX=list(UV2WithoutNA), FUN=stat)
uv1total <- tapply(AVWithoutNA, INDEX=list(UV1WithoutNA), FUN=stat)
totaltotal <- match.fun(stat)(AVWithoutNA)
subresult <- rbind(subresult, Total = uv2total)
subresult <- cbind(subresult, Total = c(uv1total, totaltotal))
subresult<-cbind(statistics = statname, round(subresult,2))
allresults[[i]] <- subresult
}
tab <- do.call(interleave,allresults)
}
#your stuff####
testdatenSPSS <- read_spss("testSPSS.sav", enc="UTF-8", autoAttachVarLabels = TRUE,
atomic.to.fac = FALSE) #put in your data here
AV = testdatenSPSS$ualter #put in dependend variable here
UV1 = testdatenSPSS$sex #put in factor 1 here
UV2 = testdatenSPSS$vgkg #put in factor 2 here
stats <- c(mean,sd,nobs,median) #put in the statistics you want here
statnames <- c("Mean“, „SD“, „N“,“Median“) #put in the labels for the statistics your want to have in the table
#calculate the table for your stuff####
tab <- createFunctionCrossMatrix(AV, UV1, UV2, stats, statnames)
#print out the table with sjPlot####
sjt.df(as.data.frame(tab) , describe=FALSE , alternateRowColors=TRUE , title=paste("Statistiken für ", get_var_labels(AV),": ", "“,get_var_labels(UV1), “ X „, get_var_labels(UV2), sep=““) , repeatHeader=TRUE , showCommentRow=TRUE , commentString=“Datenstand: XXX“ , hideProgressBar=TRUE , showRowNames=T, stringVariable = “ „) #replace the German stuff in title and commentString with your favorite string
#end of code
Hello Daniel,
my last comment is hard to read because some blank lines were erased. Here I try it again:
#########begin of code#needed packages####
library(“sjPlot”)
library(gdata)
#definition of the function that calculates the table####
createFunctionCrossMatrix <- function (AV,
UV1 = NULL,
UV2 = NULL,
stats = NULL,
statnames = NULL)
{
AVWithoutNA <- AV[!(is.na(UV2) | is.na(UV1))]
UV1WithoutNA <- to_label(UV1)[!(is.na(UV2) | is.na(UV1))]
UV2WithoutNA <- to_label(UV2)[!(is.na(UV2) | is.na(UV1))]
allresults <- list("matrix", length(stats))
for (i in 1:length(stats)) {
stat = match.fun(stats[[i]])
statname = statnames[i]
subresult <- tapply(AVWithoutNA, INDEX=list(UV1WithoutNA, UV2WithoutNA), FUN=stat)
uv2total <- tapply(AVWithoutNA, INDEX=list(UV2WithoutNA), FUN=stat)
uv1total <- tapply(AVWithoutNA, INDEX=list(UV1WithoutNA), FUN=stat)
totaltotal <- match.fun(stat)(AVWithoutNA)
subresult <- rbind(subresult, Total = uv2total)
subresult <- cbind(subresult, Total = c(uv1total, totaltotal))
subresult<-cbind(statistics = statname, round(subresult,2))
allresults[[i]] <- subresult
}
tab <- do.call(interleave,allresults)
}
#your stuff####
testdatenSPSS <- read_spss("testSPSS.sav", enc="UTF-8", autoAttachVarLabels = TRUE,
atomic.to.fac = FALSE) #put in your data here
AV = testdatenSPSS$ualter #put in dependend variable here
UV1 = testdatenSPSS$sex #put in factor 1 here
UV2 = testdatenSPSS$vgkg #put in factor 2 here
stats <- c(mean,sd,nobs,median) #put in the statistics you want here
statnames <- c("Mean”, “SD”, “N”,”Median”) #put in the labels for the statistics your want to have in the table
#calculate the table for your stuff####
tab <- createFunctionCrossMatrix(AV, UV1, UV2, stats, statnames)
#print out the table with sjPlot####
sjt.df(as.data.frame(tab) , describe=FALSE , alternateRowColors=TRUE , title=paste("Statistiken für ", get_var_labels(AV),": ", "”,get_var_labels(UV1), ” X “, get_var_labels(UV2), sep=””) , repeatHeader=TRUE , showCommentRow=TRUE , commentString=”Datenstand: XXX” , hideProgressBar=TRUE , showRowNames=T, stringVariable = ” “) #replace the German stuff in title and commentString with your favorite string
#end of code
Sorry, this concerns `sjt.glm()` function, but I guess it will have relevance here too. I was wondering if there is any way to customise the name for categorical predictor that appears on a separate row when `group.pred=TRUE` (i.e., default specification). I tried to find in the help but have been unsuccessful so far. Thanks a lot, m.
See the sjt.lm-manual (applies to sjt.glm as well), section Automatic grouping of categorical predictors and the very last example on that page. In short: use
labelPredictorsparameter to define labels for predictors, but only for those „rows“ with data (no „header-rows“). To name variables, use theset_var_labelsandset_val_labelsfunctions from the sjmisc-package (see also this manual page…).Works great, thanks a lot for your quick response and for this excellent package too! m.
Hello Daniel,
sjt.df with describe=FALSE works very fine, thanks again. Now I’ve a special data situation. One of the values in the data frame is 2.00. Unfortunately it is print as 2 in the output-table. All other values are shown with their decimals.
The problem is solved by
format(round(variable, digits=2) , nsmall=2)
Maybe this was helpful.
Thomas
Hi,
Thanks for this great package!
I’m able to use your knitr example above, with the `r sjt.lm(…)$knitr`
But when I try to do this within a chunk „`r …. „` knitr just prints out the html instead of interpreting it. I realize this is probably a knitr issue, but if you have any suggestions that would be great.
Thanks
S
Have you seen the vignette here?
To print a HTML table with knitr, use
`r sjt.frq(efc$e42dep, no.output=TRUE)$knitr`. That should work.Thank you for your very nice post! But I do have problems to get the tables to print to the knitr document. When I try this in Studio, using this command in a script
`r sjt.frq(wjsn$NORDIC, no.output=TRUE)$knitr
I get this error message:
Error in parse(text=x, srcfile=src) : :18:1: unexpected INCOMPLETE_STRING 22: 23: sjt.xtab(wjsn$C7, wjsn$NORDIC) ^Calls: -> parse_all -> parse_all.character -> parse Execution halted
Any idea what the problem can be?
I have also tried your example …
data(efc)
„`{r eval=FALSE}
sjt.frq(efc$e42dep)
„`
sjt.frq(efc$e42dep, no.output=TRUE)$knitr
… this gives no errors, but the output document shows no table, only an almost empty web page where the only text is
data(efc) sji.viewSPSS(efc)
Any idea what the problem might be?
Jan Fredrik Hovden
Thank you for such a neat package. It provides a really nice output for reports. Therefore, I am also a bit disappointed that it does not work well with knitr, which is exactly what I would hope for from a visualization package. It, however, produces nice tables that can be copy-pasted into a word processor. I am an eternal beginner with R, but I think the problem has to do with the fact that knitr processes code chunks and text chunks differently. With the code chunks, it actually prints the output and that seems to be why sjtPlot table do not get recognized as html code. In the textual part of the R markdown, the code snippets are a direct input and then the sjtPlot results get included nicely in the knitted document. I am grateful that this work-around has been made available. Thank you for the work and if you ever have the time and will to figure out how to fix the sjtPlot results in code chunks, well, it will be awesome.
Great! Nice tables!! In R!! That’s exactly what I was looking for. As far as I can see (and tried) I can only produce (and save) one table at a time. Any tips on how I manage to produce tables and attach them to the very same (html) document so that in the end I have a big output file (like in SPSS „Outputs“)? I guess that must be easy but I cannot think how to do that.
Anyway: thanks again. The ugly outputs always scared me away from R!
Each sjt-function returns parts of the HTML-table (body, style sheet) or the complete page. See return value in the help page of each function. You can then append the contents of each table, e.g.
table1 <- sjt.xtab(x1, y1)$page.contenttable2 <- sjt.xtab(x2, y2)$page.content
and then concatenate table1 and table2 via
pasteand save the complete character string with both HTML tables as html-file.I’m getting this error:
could not find function „sji.getValueLabels“
Any idea?
That was a rather old syntax from past package versions. Please update sjPlot, and refer to the package vignettes, which contain up-to-date examples.
Daniel, Your work on sjPlot is Extreme and Awesome!! Several year ago I downloaded and found it very useful in trying to help people make a transition from SPSS to R. The last version I had installed on my system was 1.8.x.
Today I downloaded and installed 2.3.1. And must say, am completely BLOWN AWAY!! Many many Cheers to you!! Keep up the great work!
Leo
Hello,
amazed to find the sji.viewSPSS() command, but even though I activated the required library require(sjPlot) (and all other the great sj-Libraries for that matter) I get the error „could not find function „sji.viewSPSS“. Am I missing something?
Best
Function names have changed over the past versions – the function is now names „view_df()“.
Hello Daniel:
Is there any way to eliminate leading zeros from the correlation coefficients?
Best,
Ale